Molecular biology d clark (elsevier, 2005)
Bạn đang xem bản rút gọn của tài liệu. Xem và tải ngay bản đầy đủ của tài liệu tại đây (39.71 MB, 802 trang )
MOLECULAR
BIOLOGY
David Clark
Southern Illinois University
AMSTERDAM • BOSTON • HEIDELBERG • LONDON
NEW YORK • OXFORD • PARIS • SAN DIEGO
SAN FRANCISCO • SINGAPORE • SYDNEY • TOKYO
MOLECULAR
BIOLOGY
Senior Acquisitions Editor
Project Manager
Editorial Assistant
Marketing Manager
Cover Design
Composition
Cover Printer
Interior Printer
Jeremy Hayhurst
Kyle Sarofeen
Desiree Marr
Linda Beattie
Eric DeCicco
SNP Best-set Typesetter Ltd., Hong Kong
Hing Yip Printing Co., Ltd.
Hing Yip Printing Co., Ltd.
Elsevier Academic Press
30 Corporate Drive, Suite 400, Burlington, MA 01803, USA
525 B Street, Suite 1900, San Diego, California 92101-4495, USA
84 Theobald’s Road, London WC1X 8RR, UK
This book is printed on acid-free paper.
Copyright © 2005, Elsevier Inc. All rights reserved.
No part of this publication may be reproduced or transmitted in any form or by any
means, electronic or mechanical, including photocopy, recording, or any information
storage and retrieval system, without permission in writing from the publisher.
Permissions may be sought directly from Elsevier’s Science & Technology Rights Department in
Oxford, UK: phone: (+44) 1865 843830, fax: (+44) 1865 853333, e-mail:
You may also complete your request on-line via the Elsevier homepage (), by
selecting “Customer Support” and then “Obtaining Permissions.”
Library of Congress Cataloging-in-Publication Data
Application submitted.
British Library Cataloguing in Publication Data
A catalogue record for this book is available from the British Library
ISBN: 0-12-175551-7
For all information on all Elsevier Academic Press Publications
visit our Web site at www.books.elsevier.com
Printed in China
05 06 07 08 09
10
9
8
7
6
5
4
3
2
1
Dedication
This book is dedicated to Lonnie Russell who was to have been my
coauthor. A few months after we started this project together, in early
July 2001, Lonnie drowned in the Atlantic Ocean off the coast of Brazil
in a tragic accident.
Preface
This book’s subtitle, Understanding the Genetic Revolution, reflects the massive surge in our understanding of the
molecular foundations of genetics in the last fifty years. In
the next half century our understanding of how living
organisms function at the molecular level, together with
our ability to intervene, will expand in ways we are only
just beginning to perceive.
Today we now know that genes are much more than
the abstract entities proposed over a century ago by
Mendel. Genes are segments of DNA molecules, carrying
encoded information. Indeed, genes have now become
chemical reagents, to be manipulated in the test tube. In
the days of classical genetics genes represented inherited
characteristics but were themselves inviolate, rather like
atoms before the twentieth century. Today both genes and
atoms have sub-components to be tinkered with.
A full understanding of how living organisms function
includes an appreciation of how cells operate at the
molecular level. This is of vital importance to all of us as
it becomes ever more clear that molecular factors underlie many health problems and diseases. While cancer is the
“classic” case of a disease that only became understandable when its genetic basis was revealed, it is not the only
one by any means. Today the molecular aspects of medicine are expanding rapidly and it will soon be possible to
personally tailor clinical treatment by taking into account
the genetic make-up of individual patients.
Rather than attempting to summarize my view of
modern molecular biology (the book itself, I hope, accomplishes that) in a short preface, I’d like to briefly address
what this book is not. It is not intended as a reference work
for faculty or researchers but rather as a survey-oriented
textbook for upper division students in a variety of biological sub-disciplines. In particular it is intended for final
year undergraduates and beginning graduate students.
This book does not attempt to be exhaustive in its
coverage, even as a textbook. There is a second book in
this series (Biotechnology: Applying the Genetic Revolution, 2006) co-authored with Nanette Pazdernik, which
essentially picks up where this book ends. Both books, I
hope, effectively survey the foundations and applications
of modern molecular genetics.
Many, perhaps most, of the students using this book
will be well versed in the basics of modern genetics and
cell biology and so can pick and choose from the topics
covered as needed. However, others will not be so well
prepared, due in part to the continuing influx into molecular biology of students from related disciplines. For them
I’ve tried to create a book whose early chapters cover the
basics, before launching out into the depths.
Because of the continuing interest in applying molecular biology to an ever widening array of topics, I have
tried to avoid overdoing detail (depth) in favour of
breadth. This in no way minimizes the importance of the
subject matter for cell biologists but instead emphasizes
that molecular biology is applicable to more than just
human medicine and health. The genetic revolution has
also greatly impacted other important areas such as agriculture, veterinary medicine, animal behaviour, evolution,
and microbiology. Students of these, and related disciplines, all need to understand molecular biology at some
level.
Finally there are no references or extra reading at the
ends of the chapters, for two reasons. My own cross-questioning has revealed that neither myself nor most of my
colleagues and students have ever actually used such textbook references just as we rarely watch the extra material
on DVDs providing actors’ insights, extra scenes, and outtakes. The student has enough to deal with in the core
material.
Secondly, anyone who wants up-to-date reference
material is far better advised to run a web search. PubMed,
Google Scholar and Scirus.com are good choices.
Feedback (hopefully positive!) is welcome.
David Clark, Carbondale, Illinois, January 2005
Acknowledgements
I would like to thank the following individuals for their
help in providing information, suggestions for improvement and encouragement: Laurie Achenbach, Rubina
Ahsan, Phil Cunningham, Michelle McGehee, Donna
Mueller, Dan Nickrent, Joan Slonczewski. Especial thanks
go to Nanette Pazdernik for help in editing many of the
chapters and to Karen Fiorino for creating most of the
artwork.
vii
Introduction
Molecular Genetics Is
Driving the Biotechnology
Revolution
Although the breeding of plants and animals goes back
thousands of years, only in the last couple of centuries has
genetics emerged as a field of scientific study. Classical
genetics emerged in the 1800s when the inheritance patterns of such things as hair or eye color were examined
and when Gregor Mendel performed his famous experiments on pea plants. Techniques revealing how the inherited characteristics that we observe daily are linked to
their underlying biochemical causes have only been developed since World War II. The resulting revelation of the
molecular basis of inheritance has resulted in the increasing use of the term “molecular.” Often the term “molecular biology” refers to the biology of those molecules
related to genes, gene products and heredity—in other
words, the term molecular biology is often substituted for
the perhaps more appropriate term, molecular genetics. A
more broad-minded definition of molecular biology
includes all aspects of the study of life from a molecular
perspective. Although the molecular details of muscle
operation or plant pigment synthesis could be included
under this definition, in practice, textbooks are limited in
length. In consequence, this book is largely devoted to the
molecular aspects of the storage and transmission of biological (i.e., genetic) information.
Although there is great diversity in the structures and
lifestyles of living organisms, viewing life at the molecular
level emphasizes the inherent unity of life processes.
Perhaps it is this emergent unity, rather than the use of
sophisticated molecular techniques, that justifies molecular biology as a discipline in its own right. Instead of an
ever-expanding hodge-podge of methods for analyzing
different organisms in more and more detail, what has
been emerged from molecular analysis is an underlying
theme of information transmission that applies to all life
forms despite their outward differences.
Society is in the midst of two scientific revolutions.
One is in the realm of technology of information, or computers, and the other in molecular biology. Both are
related to the handling of large amounts of encoded information. In one case the information is man made, or at
any rate man-encoded, and the mechanisms are artificial;
the other case deals with the genetic information that
underlies life. Biology has reached the point where the
genes that control the makeup and functioning of all living
creatures are being analyzed at the molecular level and
can be altered by genetic engineering. In fact, managing
viii
and analyzing the vast mass of genetic information constantly emerging from experimentation requires the use
of sophisticated software and powerful computers. The
emerging information revolution rivals the industrial revolution in its importance, and the consequences of today’s
findings are already changing human lives and will continue to alter the lives of future generations. Data is accumulating about the molecules of inheritance and how they
are controlled and expressed at an ever faster and faster
pace. This is largely due to improved techniques, such as
PCR (polymerase chain reaction; see Ch. 23) and DNA
(deoxyribonucleic acid) arrays (see Ch. 25). In particular,
methods have recently been developed for the rapid,
simultaneous and automated analysis of multiple samples
and/or multiple genes.
One major impact of molecular biology is in the realm
of human health. The almost complete sequence of the
DNA molecules comprising the human genome was
revealed in the year 2003. So, in theory, science has available all of the genetic information needed to make a
human being. However, the function of most of a human’s
approximately 35,000 genes remains a mystery. Still more
complex is the way in which the expression of these genes
is controlled and coordinated. Inherited diseases are due
to defective versions of certain genes or to chromosomal
abnormalities. To understand why defective genes cause
problems, it is important to investigate the normal roles
of these genes. As all disease has a genetic component, the
present trend is to redefine physical and mental health
from a genetic perspective. Even the course of an infectious disease depends to a significant extent on built-in
host responses, which are determined by host genes. For
example, humans with certain genetic constitutions are at
much greater risk than others of getting SARS, even
though this is an emerging disease that only entered the
human population in the last few years. The potential is
present to improve health and to increase human and
animal life spans by preventing disease and slowing the
aging process. Clinical medicine is changing rapidly to
incorporate these new findings.
The other main arena where biotechnology will have
a massive impact is agriculture. New varieties of genetically engineered plants and animals have already been
made and some are in agricultural use. Animals and plants
used as human food sources are being engineered to adapt
them to conditions which were previously unfavorable.
Farm animals that are resistant to disease and crop plants
that are resistant to pests are being developed in order to
increase yields and reduce costs. The impact of these
genetically modified organisms on other species and on
the environment is presently a controversial issue.
Table of Contents
CHAPTER 1
CHAPTER 2
CHAPTER 3
CHAPTER 4
CHAPTER 5
CHAPTER 6
CHAPTER 7
CHAPTER 8
CHAPTER 9
CHAPTER 10
CHAPTER 11
CHAPTER 12
CHAPTER 13
CHAPTER 14
CHAPTER 15
CHAPTER 16
CHAPTER 17
CHAPTER 18
CHAPTER 19
CHAPTER 20
CHAPTER 21
CHAPTER 22
CHAPTER 23
CHAPTER 24
CHAPTER 25
CHAPTER 26
Basic Genetics 1
Cells and Organisms 21
DNA, RNA and Protein 51
Genes, Genomes and DNA 75
Cell Division and DNA Replication 103
Transcription of Genes 132
Protein Structure and Function 154
Protein Synthesis 197
Regulation of Transcription in Prokaryotes 234
Regulation of Transcription in Eukaryotes 262
Regulation at the RNA Level 281
Processing of RNA 302
Mutations 333
Recombination and Repair 368
Mobile DNA 396
Plasmids 425
Viruses 453
Bacterial Genetics 484
Diversity of Lower Eukaryotes 508
Molecular Evolution 533
Nucleic Acids: Isolation, Purification, Detection, and
Hybridization 567
Recombinant DNA Technology 599
The Polymerase Chain Reaction 634
Genomics and DNA Sequencing 662
Analysis of Gene Expression 693
Proteomics: The Global Analysis of Proteins 717
Glossary 745
Index 771
ix
Detailed Contents
CHAPTER 1 Basic Genetics
Gregor Mendel Was the Father of
Classical Genetics
Genes Determine Each Step in Biochemical
Pathways
Mutants Result from Alterations in Genes
Phenotypes and Genotypes
Chromosomes Are Long, Thin Molecules
That Carry Genes
Different Organisms may Have Different
Numbers of Chromosomes
Dominant and Recessive Alleles
Partial Dominance, Co-Dominance,
Penetrance and Modifier Genes
Genes from Both Parents Are Mixed by
Sexual Reproduction
Sex Determination and Sex-Linked
Characteristics
Neighboring Genes Are Linked during
Inheritance
Recombination during Meiosis Ensures
Genetic Diversity
Escherichia coli Is a Model for Bacterial
Genetics
CHAPTER 2 Cells and Organisms
What Is Life?
Living Creatures Are Made of Cells
Essential Properties of a Living Cell
Prokaryotic Cells Lack a Nucleus
Eubacteria and Archaebacteria Are
Genetically Distinct
Bacteria Were Used for Fundamental Studies
of Cell Function
Escherichia coli (E. coli) Is a Model Bacterium
Where Are Bacteria Found in Nature?
Some Bacteria Cause Infectious Disease, but
Most Are Beneficial
Eukaryotic Cells Are Sub-Divided into
Compartments
The Diversity of Eukaryotes
Eukaryotes Possess Two Basic Cell Lineages
Organisms Are Classified
x
1
2
3
4
5
6
7
8
9
11
13
Some Widely Studied Organisms Serve as
Models
Yeast Is a Widely Studied Single-Celled
Eukaryote
A Roundworm and a Fly are Model
Multicellular Animals
Zebrafish are used to Study Vertebrate
Development
Mouse and Man
Arabidopsis Serves as a Model for Plants
Haploidy, Diploidy and the Eukaryote Cell
Cycle
Viruses Are Not Living Cells
Bacterial Viruses Infect Bacteria
Human Viral Diseases Are Common
A Variety of Subcellular Genetic Entities Exist
16
17
21
22
23
23
27
28
29
31
32
34
34
36
36
38
40
41
42
44
44
45
46
47
48
49
CHAPTER 3 DNA, RNA and
Protein
15
40
Nucleic Acid Molecules Carry Genetic
Information
Chemical Structure of Nucleic Acids
DNA and RNA Each Have Four Bases
Nucleosides Are Bases Plus Sugars;
Nucleotides Are Nucleosides Plus Phosphate
Double Stranded DNA Forms a Double Helix
Base Pairs are Held Together by Hydrogen
Bonds
Complementary Strands Reveal the Secret of
Heredity
Constituents of Chromosomes
The Central Dogma Outlines the Flow of
Genetic Information
Ribosomes Read the Genetic Code
The Genetic Code Dictates the Amino Acid
Sequence of Proteins
Various Classes of RNA Have Different
Functions
Proteins, Made of Amino Acids, Carry Out
Many Cell Functions
The Structure of Proteins Has Four Levels of
Organization
Proteins Vary in Their Biological Roles
51
52
52
54
55
56
57
59
60
63
65
67
69
70
71
73
Detailed Contents
CHAPTER 4 Genes, Genomes and
DNA
History of DNA as the Genetic Material
How Much Genetic Information Is Necessary
to Maintain Life?
Non-Coding DNA
Coding DNA May Be Present within
Non-coding DNA
Repeated Sequences Are a Feature of DNA in
Higher Organisms
Satellite DNA Is Non-coding DNA in the Form
of Tandem Repeats
Minisatellites and VNTRs
Origin of Selfish DNA and Junk DNA
Palindromes, Inverted Repeats and Stem and
Loop Structures
Multiple A-Tracts Cause DNA to Bend
Supercoiling is Necessary for Packaging of
Bacterial DNA
Topoisomerases and DNA Gyrase
Catenated and Knotted DNA Must Be
Corrected
Local Supercoiling
Supercoiling Affects DNA Structure
Alternative Helical Structures of DNA Occur
Histones Package DNA in Eukaryotes
Further Levels of DNA Packaging in
Eukaryotes
Melting Separates DNA Strands; Cooling
Anneals Them
75
76
78
78
80
81
83
84
84
86
87
88
89
91
91
91
92
95
96
100
CHAPTER 5 Cell Division and
DNA Replication
Cell Division and Reproduction Are Not Always
Identical
DNA Replication Is a Two-Stage Process
Occurring at the Replication Fork
Supercoiling Causes Problems for Replication
Strand Separation Precedes DNA Synthesis
Properties of DNA Polymerase
Polymerization of Nucleotides
Supplying the Precursors for DNA Synthesis
DNA Polymerase Elongates DNA Strands
The Complete Replication Fork Is Complex
Discontinuous Synthesis of DNA Requires
a Primosome
Completing the Lagging Strand
103
104
104
105
107
107
109
109
111
112
114
116
Chromosome Replication Initiates at oriC
DNA Methylation and Attachment to the
Membrane Control Initiation of Replication
Chromosome Replication Terminates at terC
Disentangling the Daughter Chromosomes
Cell Division in Bacteria Occurs after
Replication of Chromosomes
How Long Does It Take for Bacteria to
Replicate?
The Concept of the Replicon
Replicating Linear DNA in Eukaryotes
Eukaryotic Chromosomes Have Multiple Origins
Synthesis of Eukaryotic DNA
Cell Division in Higher Organisms
xi
118
120
121
122
124
124
125
126
129
130
130
CHAPTER 6 Transcription of Genes
132
Genes are Expressed by Making RNA
Short Segments of the Chromosome Are
Turned into Messages
Terminology: Cistrons, Coding Sequences and
Open Reading Frames
How Is the Beginning of a Gene Recognized?
Manufacturing the Message
RNA Polymerase Knows Where to Stop
How Does the Cell Know Which Genes to
Turn On?
What Activates the Activator?
Negative Regulation Results from the Action
of Repressors
Many Regulator Proteins Bind Small Molecules
and Change Shape
Transcription in Eukaryotes Is More Complex
Transcription of rRNA and tRNA in Eukaryotes
Transcription of Protein-Encoding Genes in
Eukaryotes
Upstream Elements Increase the Efficiency
of RNA Polymerase II Binding
Enhancers Control Transcription at a Distance
133
134
134
135
137
138
140
141
143
144
145
146
148
151
152
CHAPTER 7 Protein Structure and
Function
154
Proteins Are Formed from Amino Acids
Formation of Polypeptide Chains
Twenty Amino Acids Form Biological
Polypeptides
Amino Acids Show Asymmetry around the
Alpha-carbon
155
155
155
158
xii
Detailed Contents
The Structure of Proteins Reflects Four Levels
of Organization
The Secondary Structure of Proteins Relies on
Hydrogen Bonds
The Tertiary Structure of Proteins
A Variety of Forces Maintain the 3-D Structure
of Proteins
Cysteine Forms Disulfide Bonds
Multiple Folding Domains in Larger Proteins
Quaternary Structure of Proteins
Higher Level Assemblies and Self-Assembly
Cofactors and Metal Ions Are Often Associated
with Proteins
Nucleoproteins, Lipoproteins and Glycoproteins
Are Conjugated Proteins
Proteins Serve Numerous Cellular Functions
Protein Machines
Enzymes Catalyze Metabolic Reactions
Enzymes Have Varying Specificities
Lock and Key and Induced Fit Models Describe
Substrate Binding
Enzymes Are Named and Classified According
to the Substrate
Enzymes Act by Lowering the Energy
of Activation
The Rate of Enzyme Reactions
Substrate Analogs and Enzyme Inhibitors Act
at the Active Site
Enzymes May Be Directly Regulated
Allosteric Enzymes Are Affected by Signal
Molecules
Enzymes May Be Controlled by Chemical
Modification
Binding of Proteins to DNA Occurs in Several
Different Ways
Denaturation of Proteins
CHAPTER 8 Protein Synthesis
Protein Synthesis Follows a Plan
Proteins Are Gene Products
Decoding the Genetic Code
Transfer RNA Forms a Flat Cloverleaf Shape
and a Folded “L” Shape
Modified Bases Are Present in Transfer RNA
Some tRNA Molecules Read More Than
One Codon
160
160
163
165
166
166
167
169
169
172
174
177
177
179
181
181
182
184
184
187
187
189
190
194
197
Charging the tRNA with the Amino Acid
The Ribosome: The Cell’s Decoding Machine
Three Possible Reading Frames Exist
The Start Codon Is Chosen
The Initiation Complexes Must Be Assembled
The tRNA Occupies Three Sites During
Elongation of the Polypeptide
Termination of Protein Synthesis Requires
Release Factors
Several Ribosomes Usually Read the Same
Message at Once
Bacterial Messenger RNA Can Code for
Several Proteins
Transcription and Translation Are Coupled in
Bacteria
Some Ribosomes Become Stalled and Are
Rescued
Differences between Eukaryotic and
Prokaryotic Protein Synthesis
Initiation of Protein Synthesis in Eukaryotes
Protein Synthesis Is Halted When Resources
Are Scarce
A Signal Sequence Marks a Protein for Export
from the Cell
Molecular Chaperones Oversee Protein
Folding
Protein Synthesis Occurs in Mitochondria and
Chloroplasts
Proteins Are Imported into Mitochondria and
Chloroplasts by Translocases
Mistranslation Usually Results in Mistakes in
Protein Synthesis
The Genetic Code Is Not “Universal”
Unusual Amino Acids are Made in Proteins by
Post-Translational Modifications
Selenocysteine: The 21st Amino Acid
Pyrrolysine: The 22nd Amino Acid
Many Antibiotics Work by Inhibiting Protein
Synthesis
Degradation of Proteins
198
198
199
CHAPTER 9 Regulation of
200
201
Gene Regulation Ensures a Physiological
Response
Regulation at the Level of Transcription
Involves Several Steps
202
Transcription in
Prokaryotes
204
204
208
210
211
211
213
214
215
216
217
218
218
221
221
224
225
226
226
227
227
227
228
230
231
234
235
236
Detailed Contents
Alternative Sigma Factors in Prokaryotes
Recognize Different Sets of Genes
Heat Shock Sigma Factors in Prokaryotes Are
Regulated by Temperature
Cascades of Alternative Sigma Factors Occur
in Bacillus Spore Formation
Anti-sigma Factors Inactivate Sigma;
Anti-anti-sigma Factors Free It to Act
Activators and Repressors Participate in
Positive and Negative Regulation
The Operon Model of Gene Regulation
Some Proteins May Act as Both Repressors
and Activators
Nature of the Signal Molecule
Activators and Repressors May Be Covalently
Modified
Two-Component Regulatory Systems
Phosphorelay Systems
Specific Versus Global Control
Crp Protein Is an Example of a Global
Control Protein
Accessory Factors and Nucleoid Binding
Proteins
Action at a Distance and DNA Looping
Anti-termination as a Control Mechanism
238
238
Transcriptional Regulation in Eukaryotes Is
More Complex Than in Prokaryotes
Specific Transcription Factors Regulate Protein
Encoding Genes
The Mediator Complex Transmits Information
to RNA Polymerase
Enhancers and Insulator Sequences Segregate
DNA Functionally
Matrix Attachment Regions Allow DNA
Looping
Negative Regulation of Transcription Occurs
in Eukaryotes
Heterochromatin Causes Difficulty for Access
to DNA in Eukaryotes
Methylation of DNA in Eukaryotes Controls
Gene Expression
Silencing of Genes Is Caused by DNA
Methylation
275
277
239
242
243
244
246
248
252
253
254
254
255
256
257
258
CHAPTER 10 Regulation of
Transcription in
Eukaryotes
Genetic Imprinting in Eukaryotes Has Its
Basis in DNA Methylation Patterns
X-chromosome Inactivation Occurs in Female
XX Animals
xiii
262
CHAPTER 11 Regulation at the
RNA Level
Regulation at the Level of RNA
Binding of Proteins to mRNA Controls
The Rate of Degradation
Some mRNA Molecules Must Be Cleaved
Before Translation
Some Regulatory Proteins May Cause
Translational Repression
Some Regulatory Proteins Can Activate
Translation
Translation May Be Regulated by
Antisense RNA
Regulation of Translation by Alterations to
the Ribosome
RNA Interference (RNAi)
Amplification and Spread of RNAi
Experimental Administration of siRNA
PTGS in Plants and Quelling in Fungi
Micro RNA—A Class of Small
Regulatory RNA
Premature Termination Causes Attenuation of
RNA Transcription
Riboswitches—RNA Acting Directly as a
Control Mechanism
281
282
282
283
284
287
288
290
291
292
293
294
295
297
299
263
264
264
265
268
269
270
273
275
CHAPTER 12 Processing of RNA
RNA is Processed in Several Ways
Coding and Non-Coding RNA
Processing of Ribosomal and Transfer RNA
Eukaryotic Messenger RNA Contains a Cap
and Tail
Capping is the First Step in Maturation of
mRNA
A Poly(A) Tail is Added to Eukaryotic mRNA
Introns are Removed from RNA by Splicing
Different Classes of Intron Show Different
Splicing Mechanisms
Alternative Splicing Produces Multiple Forms of
RNA
302
303
304
305
305
306
308
310
314
315
xiv
Detailed Contents
Inteins and Protein Splicing
Base Modification of rRNA Requires Guide
RNA
RNA Editing Involves Altering the Base
Sequence
Transport of RNA out of the Nucleus
Degradation of mRNA
Nonsense Mediated Decay of mRNA
CHAPTER 13 Mutations
Mutations Alter the DNA Sequence
The Major Types of Mutation
Base Substitution Mutations
Missense Mutations May Have Major or
Minor Effects
Nonsense Mutations Cause Premature
Polypeptide Chain Termination
Deletion Mutations Result in Shortened or
Absent Proteins
Insertion Mutations Commonly Disrupt
Existing Genes
Frameshift Mutations Sometimes Produce
Abnormal Proteins
DNA Rearrangements Include Inversions,
Translocations, and Duplications
Phase Variation Is Due to Reversible DNA
Alterations
Silent Mutations Do Not Alter the Phenotype
Chemical Mutagens Damage DNA
Radiation Causes Mutations
Spontaneous Mutations Can Be Caused by
DNA Polymerase Errors
Mutations Can Result from Mispairing and
Recombination
Spontaneous Mutation Can Be the Result of
Tautomerization
Spontaneous Mutation Can Be Caused by
Inherent Chemical Instability
Mutations Occur More Frequently at Hot Spots
How Often Do Mutations Occur?
Reversions Are Genetic Alterations That
Change the Phenotype Back to Wild-type
Reversion Can Occur by Compensatory
Changes in Other Genes
Altered Decoding by Transfer RNA May
Cause Suppression
Mutagenic Chemicals Can Be Detected by
Reversion
318
322
324
327
327
328
333
334
335
336
336
338
340
341
343
343
345
346
348
350
351
353
353
353
355
358
359
361
362
363
Experimental Isolation of Mutations
In Vivo versus In Vitro Mutagenesis
Site-Directed Mutagenesis
364
365
366
CHAPTER 14 Recombination and
Repair
Overview of Recombination
Molecular Basis of Homologous Recombination
Single-Strand Invasion and Chi Sites
Site-Specific Recombination
Recombination in Higher Organisms
Overview of DNA Repair
DNA Mismatch Repair System
General Excision Repair System
DNA Repair by Excision of Specific Bases
Specialized DNA Repair Mechanisms
Photoreactivation Cleaves Thymine Dimers
Transcriptional Coupling of Repair
Repair by Recombination
SOS Error Prone Repair in Bacteria
Repair in Eukaryotes
Double-Strand Repair in Eukaryotes
Gene Conversion
CHAPTER 15 Mobile DNA
368
369
370
371
373
376
378
379
381
383
384
387
387
388
388
391
392
392
396
Sub-Cellular Genetic Elements as Gene
Creatures
Most Mobile DNA Consists of Transposable
Elements
The Essential Parts of a Transposon
Insertion Sequences—the Simplest Transposons
Movement by Conservative Transposition
Complex Transposons Move by Replicative
Transposition
Replicative and Conservative Transposition are
Related
Composite Transposons
Transposition may Rearrange Host DNA
Transposons in Higher Life Forms
Retro-Elements Make an RNA Copy
Repetitive DNA of Mammals
Retro-Insertion of Host-Derived DNA
Retrons Encode Bacterial Reverse Transcriptase
406
406
408
410
412
414
415
416
The Multitude of Transposable Elements
417
397
397
398
400
401
402
Detailed Contents
Bacteriophage Mu is a Transposon
Conjugative Transposons
Integrons Collect Genes for Transposons
Junk DNA and Selfish DNA
Homing Introns
CHAPTER 16 Plasmids
Plasmids as Replicons
General Properties of Plasmids
Plasmid Families and Incompatibility
Occasional Plasmids are Linear or Made of
RNA
Plasmid DNA Replicates by Two Alternative
Methods
Control of Copy Number by Antisense RNA
Plasmid Addiction and Host Killing Functions
Many Plasmids Help their Host Cells
Antibiotic Resistance Plasmids
Mechanism of Antibiotic Resistance
Resistance to Beta-Lactam Antibiotics
Resistance to Chloramphenicol
Resistance to Aminoglycosides
Resistance to Tetracycline
Resistance to Sulfonamides and Trimethoprim
Plasmids may Provide Aggressive Characters
Most Colicins Kill by One of Two Different
Mechanisms
Bacteria are Immune to their own Colicins
Colicin Synthesis and Release
Virulence Plasmids
Ti-Plasmids are Transferred from Bacteria to
Plants
The 2 Micron Plasmid of Yeast
Certain DNA Molecules may Behave as
Viruses or Plasmids
417
420
420
422
423
425
426
427
428
428
430
432
435
436
436
438
438
439
440
441
442
442
444
445
446
446
447
450
451
DNA Viruses of Higher Organisms
Viruses with RNA Genomes Have Very Few
Genes
Bacterial RNA Viruses
Double Stranded RNA Viruses of Animals
Positive-Stranded RNA Viruses Make
Polyproteins
Strategy of Negative-Strand RNA Viruses
Plant RNA Viruses
Retroviruses Use both RNA and DNA
Genome of the Retrovirus
Subviral Infectious Agents
Satellite Viruses
Viroids are Naked Molecules of Infectious RNA
Prions are Infectious Proteins
CHAPTER 18 Bacterial Genetics
Reproduction versus Gene Transfer
Fate of the Incoming DNA after Uptake
Transformation is Gene Transfer by Naked DNA
Transformation as Proof that DNA is the
Genetic Material
Transformation in Nature
Gene Transfer by Virus—Transduction
Generalized Transduction
Specialized Transduction
Transfer of Plasmids between Bacteria
Transfer of Chromosomal Genes Requires
Plasmid Integration
Gene Transfer among Gram-Positive Bacteria
Archaebacterial Genetics
Whole Genome Sequencing
Viruses are Infectious Packages of Genetic
Information
Life Cycle of a Virus
Bacterial Viruses are Known as Bacteriophage
Lysogeny or Latency by Integration
The Great Diversity of Viruses
Small Single-Stranded DNA Viruses of Bacteria
Complex Bacterial Viruses with Double
Stranded DNA
453
454
455
458
460
462
463
465
466
467
469
469
469
470
470
472
477
477
479
480
481
484
485
485
487
488
491
493
493
494
495
496
501
504
506
CHAPTER 19 Diversity of
Lower Eukaryotes
CHAPTER 17 Viruses
xv
Origin of the Eukaryotes by Symbiosis
The Genomes of Mitochondria and Chloroplasts
Primary and Secondary Endosymbiosis
Is Malaria Really a Plant?
Symbiosis: Parasitism versus Mutualism
Bacerial Endosymbionts of Killer Paramecium
Is Buchnera an Organelle or a Bacterium?
Ciliates have Two Types of Nucleus
Trypanosomes Vary Surface Proteins to Outwit
the Immune System
508
509
510
511
512
515
515
517
517
520
xvi
Detailed Contents
Mating Type Determination in Yeast
Multi-Cellular Organisms and Homeobox Genes
525
530
CHAPTER 20 Molecular Evolution
533
Getting Started—Formation of the Earth
The Early Atmosphere
Oparin’s Theory of the Origin of Life
The Miller Experiment
Polymerization of Monomers to Give
Macromolecules
Enzyme Activities of Random Proteinoids
Origin of Informational Macromolecules
Ribozymes and the RNA World
The First Cells
The Autotrophic Theory of the Origin of
Metabolism
Evolution of DNA, RNA and Protein
Sequences
Creating New Genes by Duplication
Paralogous and Orthologous Sequences
Creating New Genes by Shuffling
Different Proteins Evolve at Very Different
Rates
Molecular Clocks to Track Evolution
Ribosomal RNA—A Slowly Ticking Clock
The Archaebacteria versus the Eubacteria
DNA Sequencing and Biological Classification
Mitochondrial DNA—A Rapidly Ticking Clock
The African Eve Hypothesis
Ancient DNA from Extinct Animals
Evolving Sideways: Horizontal Gene Transfer
Problems in Estimating Horizontal Gene
Transfer
534
534
535
536
538
539
540
540
542
544
545
547
549
550
550
552
552
554
555
559
560
562
564
565
CHAPTER 21 Nucleic Acids:
Isolation, Purification,
Detection, and
Hybridization
567
Isolation of DNA
Purification of DNA
Removal of Unwanted RNA
Gel Electrophoresis of DNA
Pulsed Field Gel Electrophoresis
Denaturing Gradient Gel Electrophoresis
568
568
569
570
572
573
Chemical Synthesis of DNA
Chemical Synthesis of Complete Genes
Peptide Nucleic Acid
Measuring the Concentration DNA and RNA
with Ultraviolet Light
Radioactive Labeling of Nucleic Acids
Detection of Radio-Labeled DNA
Fluorescence in the Detection of DNA and RNA
Chemical Tagging with Biotin or Digoxigenin
The Electron Microscope
Hybridization of DNA and RNA
Southern, Northern, and Western Blotting
Zoo Blotting
Fluorescence in Situ Hybridization (FISH)
Molecular Beacons
574
580
580
582
583
583
585
587
588
590
592
595
595
598
CHAPTER 22 Recombinant DNA
Technology
Introduction
Nucleases Cut Nucleic Acids
Restriction and Modification of DNA
Recognition of DNA by Restriction
Endonucleases
Naming of Restriction Enzymes
Cutting of DNA by Restriction Enzymes
DNA Fragments are Joined by DNA Ligase
Making a Restriction Map
Restriction Fragment Length Polymorphisms
Properties of Cloning Vectors
Multicopy Plasmid Vectors
Inserting Genes into Vectors
Detecting Insertions in Vectors
Moving Genes between Organisms: Shuttle
Vectors
Bacteriophage Lambda Vectors
Cosmid Vectors
Yeast Artificial Chromosomes
Bacterial and P1 Artificial Chromosomes
A DNA Library Is a Collection of Genes from
One organism
Screening a Library by Hybridization
Screening a Library by Immunological
Procedures
Cloning Complementary DNA Avoids Introns
Chromosome Walking
599
600
600
600
601
601
602
603
604
607
608
610
610
612
615
616
617
620
620
621
623
623
624
626
Detailed Contents
Cloning by Subtractive Hybridization
Expression Vectors
628
631
CHAPTER 23 The Polymerase Chain
Reaction
Fundamentals of the Polymerase Chain
Reaction
Cycling Through the PCR
Degenerate Primers
Inverse PCR
Adding Artificial Restriction Sites
TA Cloning by PCR
Randomly Amplified Polymorphic DNA
(RAPD)
Reverse Transcriptase PCR
Differential Display PCR
Rapid Amplification of cDNA Ends (RACE)
PCR in Genetic Engineering
Directed Mutagenesis
Engineering Deletions and Insertions by PCR
Use of PCR in Medical Diagnosis
Environmental Analysis by PCR
Rescuing DNA from Extinct Life Forms by
PCR
Realtime Fluorescent PCR
Inclusion of Molecular Beacous in PCR—
Scorpion Primers
Rolling Circle Amplification Technology
(RCAT)
634
635
638
640
641
642
643
643
646
647
649
649
651
651
652
653
654
655
656
657
CHAPTER 24 Genomics and DNA
Sequencing
Introduction to Genomics
DNA Sequencing—General Principle
The Chain Termination Method for Sequencing
DNA
DNA Polymerases for Sequencing DNA
Producing Template DNA for Sequencing
Primer Walking along a Strand of DNA
Automated Sequencing
The Emergence of DNA Chip Technology
The Oligonucleotide Array Detector
Pyrosequencing
Nanopore Detectors for DNA
Large Scale Mapping with Sequence Tags
662
663
663
663
668
668
670
670
672
672
674
676
676
Mapping of Sequence Tagged Sites
Assembling Small Genomes by Shotgun
Sequencing
Race for the Human Genome
Assembling a Genome from Large Cloned
Contigs
Assembling a Genome by Directed Shotgun
Sequencing
Survey of the Human Genome
Sequence Polymorphisms: SSLPs and SNPs
Gene Identification by Exon Trapping
Bioinformatics and Computer Analysis
xvii
677
680
680
683
683
683
686
688
690
CHAPTER 25 Analysis of Gene
Expression
Introduction
Monitoring Gene Expression
Reporter Genes for Monitoring Gene
Expression
Easily Assayable Enzymes as Reporters
Light Emission by Luciferase as a Reporter
System
Green Fluorescent Protein as a Reporter
Gene Fusions
Deletion Analysis of the Upstream Region
Locating Protein Binding Sites in the
Upstream Region
Location of the Start of Transcription by
Primer Extension
Location of the Start of Transcription by S1
Nuclease
Transcriptome Analysis
DNA Microarrays for Gene Expression
Serial Analysis of Gene Expression (SAGE)
693
694
694
694
696
696
699
699
702
702
706
707
709
709
713
CHAPTER 26 Proteomics: The
Global Analysis of
Proteins
Introduction to Proteomics
Gel Electrophoresis of Proteins
Two Dimensional PAGE of Proteins
Western Blotting of Proteins
Mass Spectrometry for Protein Identification
Protein Tagging Systems
Full-Length Proteins Used as Fusion Tags
Self Cleavable Intein Tags
717
718
719
720
722
722
726
726
729
xviii
Detailed Contents
Selection by Phage Display
Protein Interactions: The Yeast Two-Hybrid
System
Protein Interaction by Co-Immunoprecipitation
Protein Arrays
729
Metabolomics
741
732
737
741
Glossary
745
Index
771
C H A P T E R
O N E
Basic Genetics
Gregor Mendel Was the Father of Classical Genetics
Genes Determine Each Step in Biochemical Pathways
Mutants Result from Alterations in Genes
Phenotypes and Genotypes
Chromosomes Are Long, Thin Molecules That Carry Genes
Different Organisms may Have Different Numbers of Chromosomes
Dominant and Recessive Alleles
Partial Dominance, Co-Dominance, Penetrance and Modifier Genes
Genes from Both Parents Are Mixed by Sexual Reproduction
Sex Determination and Sex-Linked Characteristics
Neighboring Genes Are Linked during Inheritance
Recombination during Meiosis Ensures Genetic Diversity
Escherichia coli Is a Model for Bacterial Genetics
1
2
CHAPTER ONE • Basic Genetics
Gregor Mendel Was the Father of Classical Genetics
A century before the discovery
of the DNA double helix,
Mendel realized that
inheritance was quantized into
discrete units we now call
genes.
From very ancient times, people have vaguely realized the basic premise of heredity. It
was always a presumption that children looked like their fathers and mothers, and that
the offspring of animals and plants generally resemble their ancestors. During the 19th
century, there was great interest in how closely offspring resembled their parents. Some
early investigators measured such quantitative characters as height, weight, or crop
yield and analyzed the data statistically. However, they failed to produce any clear-cut
theory of inheritance. It is now known that certain properties of higher organisms, such
as height or skin color, are due to the combined action of many genes. Consequently,
there is a gradation or quantitative variation in such properties. Such multi-gene characteristics caused much confusion for the early geneticists and they are still difficult to
analyze, especially if more than two or three genes are involved.
The birth of modern genetics was due to the discoveries of Gregor Mendel
(1823–1884), an Augustinian monk who taught natural science to high school students
in the town of Brno in Moravia (now part of the Czech Republic). Mendel’s greatest
insight was to focus on discrete, clear-cut characters rather than measuring continuously variable properties, such as height or weight. Mendel used pea plants and studied
characteristics such as whether the seeds were smooth or wrinkled, whether the flowers
were red or white, and whether the pods were yellow or green, etc. When asked if any
particular individual inherited these characteristics from its parents, Mendel could
respond with a simple “yes” or “no,” rather than “maybe” or “partly.” Such clear-cut,
discrete characteristics are known as Mendelian characters (Fig. 1.01).
Today, scientists would attribute each of the characteristics examined by Mendel
to a single gene. Genes are units of genetic information and each gene provides the
instructions for some property of the organism in question. In addition to those genes
that affect the characteristics of the organism more or less directly, there are also many
regulatory genes.These control other genes, hence their effects on the organism are less
direct and more complex. Each gene may exist in alternative forms known as alleles,
which code for different versions of a particular inherited character (such as red versus
white flower color). The different alleles of the same gene are closely related, but have
minor chemical variations that may produce significantly different outcomes.
The overall nature of an organism is due to the sum of the effects of all of its genes
as expressed in a particular environment. The total genetic make-up of an organism is
referred to as its genome. In lower organisms such as bacteria, the genome may consist
of approximately 2,000 to 6,000 genes, whereas in higher organisms such as plants and
animals, there may be up to 50,000 genes.
Etymological Note
M
endel did not use the word “gene.” This term entered the English language
in 1911 and was derived from the German “Gen,” short for “Pangen.” This
in turn came via French and Latin from the original ancient Greek “genos,” which
means birth. “Gene” is related to such modern words as genus, origin, generate,
and genesis. In Roman times, a “genius” was a spirit representing the inborn
power of individuals.
allele One particular version of a gene
gene A unit of genetic information
genome The entire genetic information of an individual organism
Gregor Mendel Discovered the basic laws of genetics by crossing pea plants
Mendelian character Trait that is clear cut and discrete and can be unambiguously assigned to one category or another
Genes Determine Each Step in Biochemical Pathways
3
Height
d color
See
ed shape
Se
Dwarf
vs
Round
vs
Wrinkled
Tall
Green
wer color
Flo
er positio
ow
n
Fl
MENDEL’S
SEVEN
CHARACTERISTICS
Axial
Mendel chose specific
characteristics, such as those shown.
As a result he obtained definitive
answers to whether or not a
particular characteristic is inherited.
Red
vs
Terminal
vs
co
Pod lor
FIGURE 1.01 Mendelian
Characters in Peas
Yellow
vs
Green
vs
Yellow
White
shape
Pod
Inflated
vs
Constricted
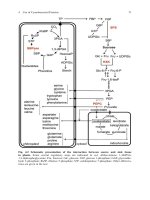
Gene
FIGURE 1.02 One Gene—
One Enzyme
A single gene determines the
presence of an enzyme which, in
turn, results in a biological
characteristic such as a red flower.
Enzyme
Precursor
Red
pigment
Genes Determine Each Step in Biochemical Pathways
Beadle and Tatum linked genes
to biochemistry by proposing
there was one gene for each
enzyme.
Much of modern molecular
biology deals with how genes
are regulated. (See Chapters
9, 10 and 11.)
Mendelian genetics was a rather abstract subject, since no one knew what genes were
actually made of, or how they operated. The first great leap forward came when biochemists demonstrated that each step in a biochemical pathway was determined by a
single gene. Each biosynthetic reaction is carried out by a specific protein known as an
enzyme. Each enzyme has the ability to mediate one particular chemical reaction and
so the one gene—one enzyme model of genetics (Fig. 1.02) was put forward by G. W.
Beadle and E. L. Tatum, who won a Nobel prize for this scheme in 1958. Since then, a
variety of exceptions to this simple scheme have been found.For example,some complex
enzymes consist of multiple subunits, each of which requires a separate gene.
A gene determining whether flowers are red or white would be responsible for a
step in the biosynthetic pathway for red pigment. If this gene were defective, no red
pigment would be made and the flowers would take the default coloration—white. It
is easy to visualize characters such as the color of flowers, pea pods or seeds in terms
of a biosynthetic pathway that makes a pigment. But what about tall versus dwarf
plants and round versus wrinkled seeds? It is difficult to interpret these in terms of a
single pathway and gene product. Indeed, these properties are affected by the action
enzyme A protein that carries out a chemical reaction
protein A polymer made from amino acids; proteins make up most of the structures in the cell and also do most of the work
4
CHAPTER ONE • Basic Genetics
Wild-type gene
Enzyme
Precursor
Red
pigment
Red
flowers (R)
No
pigment
White
flowers (r)
FIGURE 1.03 Wild-type
and Mutant Genes
If red flowers are found normally in
the wild, the “red” version of the
gene is called the wild-type allele.
Mutation of the wild-type gene may
alter the function of the enzyme so
ultimately affecting a visible
characteristic. Here, no pigment is
made and the flower is no longer
red.
Mutant gene
No
enzyme made
Precursor
of many proteins. However, as will be discussed in detail later, certain proteins control
the expression of genes rather than acting as enzymes. Some of these regulatory proteins control just one or a few genes whereas others control large numbers of genes.
Thus a defective regulatory protein may affect the levels of many other proteins.
Modern analysis has shown that some types of dwarfism are due to defects in a single
regulatory protein that controls many genes affecting growth. If the concept of “one
gene—one enzyme” is broadened to “one gene—one protein,” it still applies in most
cases. [There are of course exceptions. Perhaps the most important is that in higher
organisms multiple related proteins may sometimes be made from the same gene by
alternative patterns of splicing at the RNA level, as discussed in Chapter 12.]
Mutants Result from Alterations in Genes
Genetics has been culturally
influenced by idealized notions
of a perfect “natural” or
“original” state. Mutations tend
to be viewed as defects
relative to this.
Consider a simple pathway in which red pigment is made from its precursor in a
single step. When everything is working properly, the flowers shown in Figure 1.02
will be red and will match thousands of other red flowers growing in the wild. If the
gene for flower color is altered so as to prevent the gene from functioning properly, one
may find a plant with white flowers. Such genetic alterations are known as mutations.
The white version of the flower color gene is defective and is a mutant allele. The properly functioning red version of this gene is referred to as the wild-type allele (Fig. 1.03).
As the name implies, the wild-type is supposedly the original version as found in the
wild, before domestication and/or mutation altered the beauties of nature. In fact, there
are frequent genetic variants in wild populations and it is not always obvious which
version of a gene should be regarded as the true wild-type. Generally, the wild-type is
taken as the form that is common and shows adaptation to the environment.
Geneticists often refer to the red allele as “R” and the white allele as “r” (not
“W”). Although this may seem a strange way to designate the color white, the idea is
mutation An alteration in the genetic information carried by a gene
regulatory protein A protein that regulates the expression of a gene or the activity of another protein
wild-type The original or “natural” version of a gene or organism
Phenotypes and Genotypes
FIGURE 1.04 Three Step
Biochemical Pathway
In this scenario, genes A, B, and C
are all needed to make the red
pigment required to produce a red
flower. If any precursor is missing
due to a defective gene, the
pigment will not be made and the
flower will be white.
Precursor P
Gene A
Gene B
Gene C
Enzyme A
Enzyme B
Enzyme C
Precursor Q
Precursor R
5
Red
pigment
that the r-allele is merely a defective version of the gene for red pigment. The r-allele
is NOT a separate gene for making white color. In our hypothetical example, there is
no enzyme that makes white pigment; there is simply a failure to make red pigment.
Originally it was thought that each enzyme was either present or absent; that is, there
were two alleles corresponding to Mendel’s “yes” and “no” situations. In fact, things
are often more complicated. An enzyme may be only partially active or even be hyperactive or have an altered activity and genes may actually have dozens of alleles, matters
to be discussed later.A mutant allele that results in the complete absence of the protein
is known as a null allele. [More strictly, a null allele is one that results in complete
absence of the gene product. This includes the absence of RNA (rather than protein)
in the case of those genes where RNA is the final gene product (e.g. ribosomal RNA,
transfer RNA etc)—see Chapter 3].
Phenotypes and Genotypes
Classical genetic analysis
involves deducing the state of
the genes by observing the
outward properties of the
organism.
In real life, most biochemical pathways have several steps, not just one. To illustrate
this, extend the pathway that makes red pigment so it has three steps and three genes,
called A, B, and C. If any of these three genes is defective, the corresponding enzyme
will be missing, the red pigment will not be made, and the flowers will be white. Thus
mutations in any of the three genes will have the same effect on the outward appearance of the flowers. Only if all three genes are intact will the pathway succeed in making
its final product (Fig. 1.04).
Outward characteristics—the flower color—are referred to as the phenotype and
the genetic make-up as the genotype. Obviously, the phenotype “white flowers” may
be due to several possible genotypes, including defects in gene A, B, or C, or in genes
not mentioned here that are responsible for producing precursor P in the first place.
If white flowers are seen, only further analysis will show which gene or genes are defective. This might involve assaying the biochemical reactions, measuring the build-up of
pathway intermediates (such as P or Q in the example) or mapping the genetic defects
to locate them in a particular gene(s).
If gene A is defective, it no longer matters whether gene B or gene C are functional or not (at least as far as production of our red pigment is concerned; some genes
affect multiple pathways, a possibility not considered in this analysis). A defect near
the beginning of a pathway will make the later reactions irrelevant. This is known in
genetic terminology as epistasis. Gene A is epistatic on gene B and gene C; that is, it
masks the effects of these genes. Similarly, gene B is epistatic on gene C. From a practical viewpoint, this means that a researcher cannot tell if genes B or C are defective
or not, when there is already a defect in gene A.
epistasis When a mutation in one gene masks the effect of alterations in another gene
genotype The genetic make-up of an organism
null allele Mutant version of a gene which completely lacks any activity
phenotype The visible or measurable effect of the genotype
6
CHAPTER ONE • Basic Genetics
Chromosome
A
B
C D
E
F
GH
I
J
Genes
FIGURE 1.05
Genes Arranged along a Chromosome
Although a chromosome is a complex three-dimensional structure, the genes on a chromosome are in
linear order and can be represented by segments of a bar, as shown here. Genes are often given
alphabetical designations in genetic diagrams.
FIGURE 1.06 Circular DNA
from a Bacterium
Hand-tinted transmission electron
micrograph (TEM) of circular
bacterial DNA. This figure actually
shows a small plasmid, rather than
a full-size chromosome. The doublestranded DNA is yellow. An
individual gene has been mapped
by using an RNA copy of the gene.
The RNA base pairs to one strand
of the DNA forming a DNA/RNA
hybrid (red). The other strand of the
DNA forms a single-stranded loop,
known as an “R-loop” (blue).
Magnification: ¥28,600. Courtesy
of: P. A. McTurk and David Parker,
Science Photo Library.
Genes are not mere
abstractions. They are
segments of DNA molecules
carrying encoded information.
Chromosomes Are Long, Thin Molecules That
Carry Genes
Genes are aligned along very long, string-like molecules called chromosomes (Fig.
1.05). Organisms such as bacteria usually fit all their genes onto a single circular chromosome (Fig. 1.06); whereas, higher, eukaryotic organisms have several chromosomes
that accommodate their much greater number of genes. Genes are often drawn on a
bar representing a chromosome (or a section of one), as shown in Figure 1.05.
One entire chromosome strand consists of a molecule of deoxyribonucleic acid,
called simply DNA (see Ch. 3). The genes of living cells are made of DNA, as are the
bacteria Primitive single-celled organisms without a nucleus and with one copy of each gene
chromosome Structure containing the genes of a cell and made of a single molecule of DNA
Different Organisms may Have Different Numbers of Chromosomes
FIGURE 1.07 Genes Match
on Each of a Pair of
Homologous Chromosomes
Higher organisms possess two
copies of each gene arranged on
pairs of homologous chromosomes.
The genes of the paired
chromosomes are matched along
their length. Although
corresponding genes match, there
may be molecular variation
between the two members of each
pair of genes.
A1
B1
C1 D1 E1
Copy 1
Chromosomes
Copy 2
A2
B2
F1
G1 H1
I1
J1
I2
J2
7
Duplicate genes
C2 D2 E2
F2
G2 H2
regions of the chromosome between the genes. In bacteria, the genes are closely
packed together, but in higher organisms such as plants and animals, the DNA between
genes comprises up to 96% of the chromosome and the functional genes only make
up around 4 to 5% of the length. [Viruses also contain genetic information and some
have genes made of DNA. Other viruses have genes made of the related molecule,
ribonucleic acid, RNA.]
In addition to the DNA, the genetic material itself, chromosomes carry a variety
of proteins that are bound to the DNA. This is especially true of the larger chromosomes of higher organisms where histone proteins are important in maintaining chromosome structure (see Ch. 4). [Bacteria also have histone-like proteins. However, these
differ significantly in both structure and function from the true histones of higher
organisms—see Chapter 9.]
Different Organisms may Have Different Numbers
of Chromosomes
Different organisms differ
greatly in the number of
genes, the number of copies of
each gene, and the
arrangement of the genes on
the DNA.
The cells of higher organisms usually contain two copies of each chromosome. Each
pair of identical chromosomes possesses copies of the same genes, arranged in the same
linear order. In Figure 1.07, identical capital letters indicate sites where alleles of the
same gene can be located on a pair of chromosomes. In fact, identical chromosomes
are not usually truly identical, as the two members of the pair often carry different
alleles of the same gene. The term homologous refers to chromosomes that carry the
same set of genes in the same sequence, although they may not necessarily carry identical alleles of each gene.
A cell or organism that possesses two homologous copies of each of its chromosomes is said to be diploid (or “2n”, where “n” refers to the number of chromosomes in
one complete set).Those that possess only a single copy of each chromosome are haploid
(or “n”).Thus humans have 2 ¥ 23 chromosomes (n = 23 and 2n = 46).Although the X and
Y sex chromosomes of animals form a pair they are not actually identical (see below).
Thus, strictly, a male mammal is not fully diploid. Even in a diploid organism, the reproductive cells, known as gametes, possess only a single copy of each chromosome and are
thus haploid. Such a single, though complete, set of chromosomes carrying one copy of
each gene from a normally diploid organism is known as its “haploid genome.”
Bacteria possess only one copy of each chromosome and are therefore haploid.
(In fact, most bacteria have only a single copy of a single chromosome, so that n = 1).
If one of the genes of a haploid organism is defective, the organism may be seriously
endangered since the damaged gene no longer contains the correct information that
the cell needs. Higher organisms generally avoid this predicament by being diploid and
having duplicate copies of each chromosome and therefore of each gene. If one copy
of the gene is defective, the other copy may produce the correct product required by
the cell. Another advantage of diploidy is that it allows recombination between two
copies of the same gene (see Ch. 14). Recombination is important in promoting the
genetic variation needed for evolution.
diploid Having two copies of each gene
haploid Having one copy of each gene
haploid genome A complete set containing a single copy of all the genes (generally used of organisms that have two or more sets of each gene)
homologous Related in sequence to an extent that implies common genetic ancestry
ribonucleic acid (RNA) Nucleic acid that differs from DNA in having ribose in place of deoxyribose and having uracil in place of thymine